In a recent publication, Fujisawa et al. [1] describe how they used Voloom to reconstruct immunofluorescently stained adjacent sections. Apart from visualizing geometric structures, this enables them to study and analyze the presence and interactions of multiple specific proteins and molecules in high resolution (using Imaris from Bitplane). This would have been very challenging in tissue blocks, thick sections, or whole mount samples.
Reconstructed brain region showing complex interactions between blood vessels (red, CD31), cancer cells (green, GFP), and astrocytes (white, GFAP)
[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]
Reconstructed CD31-positive blood vessel (red) and spot detection of pMAPK-positive cells with color-coded distance information
[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]
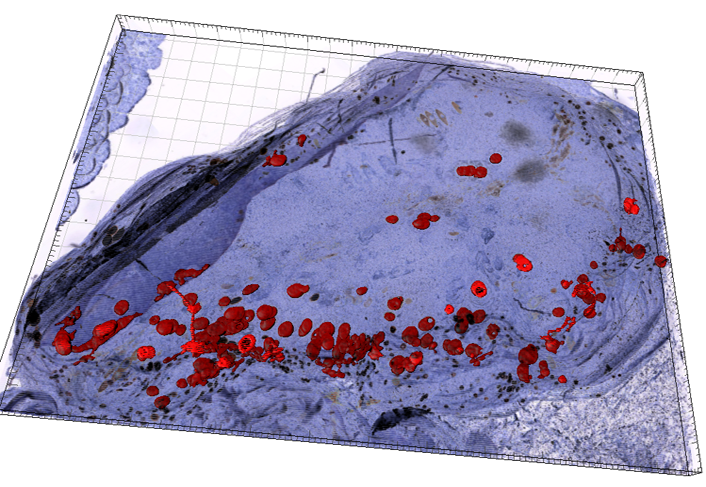
CD31 stained blood vessels in reconstructed myxofibrosarcoma
[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]
In their study they focused on high automation of the processing, staining, imaging, and alignment of sectioned paraffin embedded samples. Their method enables the visualization, analysis, as well as observation of the clear separation, localization and interaction of multiple antigens, proteins, and molecules in tissue samples with a thickness of more than 100 microns.
Thick sections (top) and reconstructed serial sections (bottom) of a mouse heart region (left) and mouse brain region (right) in comparison, showing CD31-positive vasculature (red), spot detected Ki67-positive nuclei (green), and DAPI (blue). In particular for the brain region the Ki67 signal is much more extensive in the serial sections (bottom right) than in the thick section (top right).
[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]
More details can be found in their original article:
1. Sho Fujisawa, Dmitry Yarilin, Ning Fan, Mesruh Turkekul, Ke Xu, Afsar Barlas, and Katia Manova-Todorova, "Understanding 3-Dimensional World from 2-Dimensional Immunofluorescent Adjacent Sections," Analytical Cellular Pathology, vol. 2014, Article ID 784937, 2 pages, 2014. doi:10.1155/2014/784937

![Reconstructed brain region showing complex interactions between blood vessels (red, CD31), cancer cells (green, GFP), and astrocytes (white, GFAP)[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]](https://images.squarespace-cdn.com/content/v1/54d89682e4b0a80ddb2c75f2/1429773382082-WE49O686ATGXXH9YWCTP/Reconstructed+brain+region)
![Reconstructed CD31-positive blood vessel (red) and spot detection of pMAPK-positive cells with color-coded distance information[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]](https://images.squarespace-cdn.com/content/v1/54d89682e4b0a80ddb2c75f2/1429773780111-7W0KWS77YQN25ERHYGQZ/Reconstructed+blood+vessel)
![CD31 stained blood vessels in reconstructed myxofibrosarcoma[Image courtesy of Sho Fujisawa, Memorial Sloan Kettering Cancer Center]](https://images.squarespace-cdn.com/content/v1/54d89682e4b0a80ddb2c75f2/1429773071949-TLR62EEEAFIGQQ68NZES/Blood+vessels+in+reconstructed+myxofibrosarcoma)
